Chapter 5: Diving Into The OpenLC Run Data
Generally speaking you don't need to further post-process the data obtained in an OpenLC run. There are times, however, in which you would like to analize carefully which are the response times in iteration #N, or simply, access to data which is not printed by default after a OLCCommander execution.
5.1 OpenLC Run Database Outline
OpenLC saves, by default, all the run (both raw an reduced) data on a directory in the server side. In this chapter I will try to tell you where and how access to this internal data.
As you probably already know (you have configured it), the rundata is saved in the directory stated in the OpenLC-config.xml (normally in /etc/OpenLC directory), under the tag rundbpath. The structure of this directory is outlined in figure 5.1.
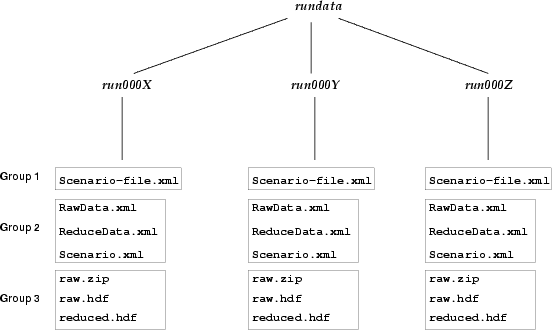
As you can see, the OpenLC run database is a two-level hierarchical directory. In the first level, there exist a directory for each run number (in the form (run%06d % runNumber)). In the second level (i.e. for each run), a number of files are created containing all the data for the current run.
These second-level files are grouped in three subsets:
- Group 1: Only one file is included here, the Scenario-file.xml, and as you may guess, it's the Scenario Definition File responsible for this run.
- Group 2: This group includes important Python
objects in XML serialization form (using the Gnosis
utilities). These objects are important to retrieve
important information of the run (for example, the number of
transactions or the actual run duration time). As the format
in which the objects are saved is XML, you can read the
values by just look at the file, which can be an important
advantage.
Three different files are present in this group:- RawData.xml
- ReduceData.xml
- Scenario.xml
- Group 3: The data coming along from the
different transactions during the run are saved in NetCDF
and HDF5 (if configured during installation) formats. As we
stated before this detailed information may be very useful
to dive into the details of the run.
Three different files are present in this group:- raw.zip: This is a ZIP file which contains the raw data (in NetCDF format) for each command separately. This files are really the original file streams where the data is saved in real-time (using the Scientific Python package) during the run.
- raw.{hdf|nc}: This is the same data than above, but consolidated in a unique file. If HDF support is there, the raw.hdf contains the arrays of command data structured in a tree. If HDF is not supported, a raw.nc is generated, with the same information, but without a tree structure and the variable names are constructed from the protocol, group and command names (for example Local-test-random-wallClock). This format is normally more difficult to manage than the tree form.
- reduced.{hdf|nc}: This file holds the reduced data resulting from a post-processing raw data process which takes place when the run is finished. The format of this data depends on the values stated in the retValues tag of the SDF file. If no HDF support, a NetCDF file is created with this same information (but with flat structure).
5.2 Browsing at the Run Raw Data
If you want to have look at the data collected by the experiment without further processing, you can start with RawData.xml. Here you have an example of the information you can find there:
<!DOCTYPE PyObject SYSTEM "PyObjects.dtd" [<PyObject class="RawData" module="OpenLC.server.RawData" id="138480564"> <attr type="string" name="hdfFilename" value="/home/falted/OpenLC/rundata/run000097/raw.hdf"></attr> <attr type="numeric" name="runid" value="97"></attr> <attr type="numeric" name="nevents_saved" value="2505"></attr> <attr type="numeric" name="nevents" value="2505"></attr> <attr type="numeric" name="elapsedTime" value="5.0204830169677734"></attr> <attr type="dict" name="filenames" id="136977668"> <entry> <key type="string" value="Local/test/random"></key> <val type="string" value="/home/falted/OpenLC/rundata/run000097/raw/Local/test/random.nc"></val> </entry> <entry> <key type="string" value="Local/test/constant"></key> <val type="string" value="/home/falted/OpenLC/rundata/run000097/raw/Local/test/constant.nc"></val> </entry> <entry> <key type="string" value="Local/test/linear"></key> <val type="string" value="/home/falted/OpenLC/rundata/run000097/raw/Local/test/linear.nc"></val> </entry> </attr> <attr type="dict" name="lastindex" id="138290436"> <entry> <key type="string" value="Local/test/random"></key> <val type="numeric" value="835"></val> </entry> <entry> <key type="string" value="Local/test/constant"></key> <val type="numeric" value="835"></val> </entry> <entry> <key type="string" value="Local/test/linear"></key> <val type="numeric" value="835"></val> </entry> </attr> <attr type="string" name="zipfile" value="/home/falted/OpenLC/rundata/run000097/raw.zip"></attr> <attr type="string" name="raw_dir" value="/home/falted/OpenLC/rundata/run000097/raw"></attr> <attr type="list" name="cmdIDs" id="137022540"> <item type="string" value="Local/test/constant"></item> <item type="string" value="Local/test/random"></item> <item type="string" value="Local/test/linear"></item> </attr> <attr type="string" name="currentDir" value="/home/falted/OpenLC/rundata/run000097"></attr> <attr type="string" name="datetime" value="Sun Jun 23 12:13:56 2002"></attr> </PyObject>
As you can see, you can get some interesting information here. If you look carefully at filenames tag, you will discover that it's a mapping (dictionary in Python jargon) between the commands in the Scenario and the NetCDF filenames. The files referenced by these filenames are package inside the raw.zip file.
In raw.hdf you can get all the raw data structured in a tree, which is well adpated to show the Scenario descrition for this run. The .hdf extension means that the file is in HDF5 format and there is a variety of software and utilities to read it. I would recommend you a couple: HDF5 tools (http://hdf.ncsa.uiuc.edu/hdf5tools.html) and HDFView (http://hdf.ncsa.uiuc.edu/hdf-java-html/hdfview/). If you don't have HDF5 support, I suggest you to have a look at the utilities ncdump and ncgen which comes with the NetCDF library or the excellent plotting tool called grace (http://plasma-gate.weizmann.ac.il/Grace/).
Next, I'll describe shortly some of these utilities.
5.2.1 HDF5 tools
There is quite a few utilities to deal with HDF5 files in HDF5 tools, but the most important ones are h5dump and h5ls. You can get instructions by passing them the flag -?. As an example of use, look at the next command and its output:
$ h5ls -r raw.hdf
/raw.hdf/Local Group
/raw.hdf/Local/test Group
/raw.hdf/Local/test/constant Group
/raw.hdf/Local/test/constant/commandNumber Dataset {835}
/raw.hdf/Local/test/constant/dataTransferred Dataset {835}
/raw.hdf/Local/test/constant/threadNumber Dataset {835}
/raw.hdf/Local/test/constant/timeSpent Dataset {835}
/raw.hdf/Local/test/constant/wallClock Dataset {835}
/raw.hdf/Local/test/linear Group
/raw.hdf/Local/test/linear/commandNumber Dataset {835}
/raw.hdf/Local/test/linear/dataTransferred Dataset {835}
/raw.hdf/Local/test/linear/threadNumber Dataset {835}
/raw.hdf/Local/test/linear/timeSpent Dataset {835}
/raw.hdf/Local/test/linear/wallClock Dataset {835}
/raw.hdf/Local/test/random Group
/raw.hdf/Local/test/random/commandNumber Dataset {835}
/raw.hdf/Local/test/random/dataTransferred Dataset {835}
/raw.hdf/Local/test/random/threadNumber Dataset {835}
/raw.hdf/Local/test/random/timeSpent Dataset {835}
/raw.hdf/Local/test/random/wallClock Dataset {835}
/raw.hdf/info Group
5.2.2 HDFView
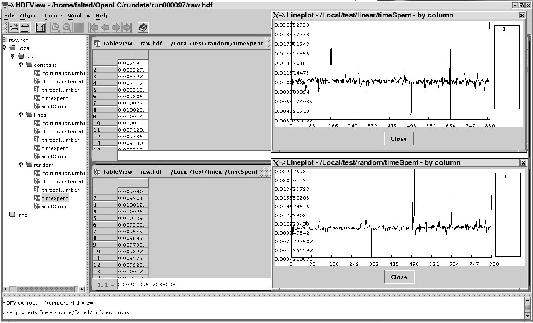
A nicer utility is HDFView. This is a Java application which lets you browse the contents of the HDF5 file, and even visualize dataset histograms. From the HDFView manual:
The HDFView is a Java-based tool for browsing and editing NCSA HDF4 and HDF5 files. HDFView allows users to browse through any HDF4 and HDF5 file; starting with a tree view of all top-level objects in an HDF file's hierarchy. HDFView allows a user to descend through the hierarchy and navigate among the file's data objects. The content of a data object is loaded only when the object is selected, providing interactive and efficient access to HDF4 and HDF5 files. HDFView editing features allow a user to create, delete, and modify the value of HDF objects and attributes.
In figure 5.2 you can see an example of an HDFView session.
5.2.3 ncdump
This utility is very useful to browse NetCDF files. From the manual:
The ncdump tool generates the CDL text representation of a netCDF file on standard output, optionally excluding some or all of the variable data in the output. The output from ncdump is intended to be acceptable as input to ncgen. Thus ncdump and ncgen can be used as inverses to transform data representation between binary and text representations. ncdump may also be used as a simple browser for netCDF data files, to display the dimension names and sizes; variable names, types, and shapes; attribute names and values; and optionally, the values of data for all variables or selected variables in a netCDF file.
5.3 The Reduced Data at a Glance
When the run is finished, the OpenLC server automatically starts a process to reduce the raw data and extract some run data statistics (following instructions in the scenario file) and saves it in files. You can find two files with the post-processed data information.
- ReduceData.xml: This file is the serialization of the ReduceData internal server class. It has a serialized hash holding mean values for all the variables for each command, group and protocol defined in the Scenario.
- reduced.hdf: Contains, depending on the
sta attribute of the retValues subtags, the next datasets
for the commands, groups and protocols:
- vds: Stands for Very Detailed Statistics and computes histograms for mean, nevents, maximum, minimum and standard deviation for bins along the wallClock variable.
- mean: Computes histogram only for mean values.
- minimal: Computes noly the mean in all the wallClock range. The result is an scalar, not a histogram.
- none: Ignore this variable in the final reduced data output.